How to manipulate Big Data using Microsoft R Server?
In this post I will demonstrate how to manipulate big data files using Mircosoft R server or MRS.
Overview
The prerequisites to reproduce the analysis in this post will be an installation of MRS or MRC (Microsoft R Client) and the data files from this very popular Kaggle competition. This post will the first of a 2 part series. I will prepare the data in this post and use it in the subsequent post for a machine learning experiment.
Understanding the data and the task
It is always in the best interest of any organization to know more about its customers. In this particular scenario, a certain company has all the tools to capture the mobile device usage behaviour of its customers through an SDK installed in its app. The company wants to predict the gender of its users basis this information. There are multiple benefits in accurately predicting this information. Typically gender is a PII and users are not willing to share it readily. By predicting the gender information the companies can gain by better segmenting its users and showing more targeted ads hence bumping up the ROI considerably on the marketing campaigns.
The data has a collection of about 6 files. As we can see below few files are pretty big to fit in to the RAM hence the need to use MRS.
library(dplyr)
data.frame(name = list.files(pattern = "csv"),
size_in_MB = file.size(list.files(pattern = "csv"))/10^6) %>%
arrange(desc(size_in_MB))
## name size_in_MB
## 1 app_events.csv 1037.267659
## 2 events.csv 195.433779
## 3 app_labels.csv 11.190003
## 4 phone_brand_device_model.csv 6.715635
## 5 gender_age_train.csv 2.366486
## 6 label_categories.csv 0.016450
Below is a breif description of information that is available:
Device Brand : Brand of the device being used by the customer
Events : Timestamp and geo-location of varous event recorded on the device
App events : List of apps involved in every event
App categories : Description of apps and its categories
Gender and Age : Gender and Age of the customer. This information will be used as label
For this experiment we will not be using the device brand as they are
not available in the English language. Also, we will use the MRS’s
RevoScaleR package when exploring the large files i.e. events and
app_events.
Data Preparation
The objective of this step is to collect various features for every user along with the gender information. By the end of this step we will target to capture the following information.
-
Gender
-
Age
-
Mean longitude of the user position
-
Mean latitude of the user position
-
Total number of apps used by every user
-
Weekday with maximum activity (based on total number of apps)
-
Day hour with maximum activity (based on total number of apps)
Lets start collecting our features by exploring various files.
Step 1 & 2: Gender & Age
# gender_age_train.csv
gender_age <- read.csv(file = "gender_age_train.csv", stringsAsFactors = TRUE)
# We will encode device_id to smaller number to save memory
map <- data.frame(device_id = gender_age$device_id, new_id = 1:nrow(gender_age))
gender_age <- merge(gender_age, map, by.x = "device_id", by.y = "device_id")
gender_age$device_id <- NULL
# Drop group columns as well as it is a combination of gender and age
gender_age$group <- NULL
head(gender_age)
## gender age new_id
## 1 M 24 6247
## 2 M 36 21595
## 3 M 29 65105
## 4 M 23 71809
## 5 F 56 67366
## 6 F 27 38986
Step 3, 4 & 5: Longitude, Latitude and Total number of Apps
We will use the RevoScaleR package to read events.csv in to an xdf
file and explore the data.
# Point to csv file
events <- "events.csv"
events.xdf <- "events.xdf"
# Import the csv into an xdf data source
events.xdf <- rxImport(events, events.xdf, stringsAsFactors = FALSE, overwrite = TRUE)
dim(events.xdf)[1]
## [1] 3252950
As we can see about 3.2M rows have been read into the xdf file. It would have been a great deal if we were to read this data to in-memory. Further, we can use various RevoScaleR functions to explore and manipulate the data.
# See top 5 rows (equivalent to head in base R)
i <- rxGetInfo(events.xdf, numRows = 5, getVarInfo = TRUE)
i$data
## event_id device_id timestamp longitude latitude
## 1 1 2.918269e+16 2016-05-01 00:55:25 121.38 31.24
## 2 2 -6.401643e+18 2016-05-01 00:54:12 103.65 30.97
## 3 3 -4.833982e+18 2016-05-01 00:08:05 106.60 29.70
## 4 4 -6.815121e+18 2016-05-01 00:06:40 104.27 23.28
## 5 5 -5.373798e+18 2016-05-01 00:07:18 115.88 28.66
We will perform the following transforms to this data:
-
Replace
device_idwithnew_idusing themapobject we created earlier -
Get the Year, Month, Day, Hour and Minute information from
timestampfor each of the events
For step a., we will use the rxMerge function
merged.xdf <- rxMerge(events.xdf,
map,
type = "inner",
matchVars = "device_id",
outFile = "merged.xdf",
overwrite = TRUE)
i <- rxGetInfo(merged.xdf, numRows = 5, getVarInfo = TRUE)
i$numRows
## [1] 1215595
The merged file has about 1.2M rows which indicates that not all
device_id have event information. We will proceed with those devices
that have the event information.
Lets move on to the next step. Adding timestamp information. We will use
the rxDataStep function to perform the transformations and write back
the changes to the same file.
rxDataStep(merged.xdf, merged.xdf, overwrite = TRUE,
transforms = list(
year = (as.POSIXlt(timestamp))$year,
month = (as.POSIXlt(timestamp))$mon,
day = factor((as.POSIXlt(timestamp))$wday, levels = 0:6,
labels = c("Su","M","Tu", "W", "Th","F", "Sa")),
hr = (as.POSIXlt(timestamp))$hour,
min = (as.POSIXlt(timestamp))$min
),
transformPackages = c("base"))
i <- rxGetInfo(merged.xdf, numRows = 5, getVarInfo = TRUE)
i$data
## event_id device_id timestamp longitude latitude new_id
## 1 11281 -9.222957e+18 2016-05-07 11:36:04 0.00 0.00 21595
## 2 12679 -9.222957e+18 2016-05-07 12:18:35 113.24 23.19 21595
## 3 49383 -9.222957e+18 2016-05-07 15:44:45 0.00 0.00 21595
## 4 131849 -9.222957e+18 2016-05-07 12:06:35 113.24 23.19 21595
## 5 205471 -9.222957e+18 2016-05-06 15:36:46 113.24 23.19 21595
## year month day hr min
## 1 116 4 Sa 11 36
## 2 116 4 Sa 12 18
## 3 116 4 Sa 15 44
## 4 116 4 Sa 12 6
## 5 116 4 F 15 36
Now we can drop device_id and timestamp variables.
merged_new.xdf <- RxXdfData("merged_new.xdf")
rxDataStep(merged.xdf,
merged_new.xdf,
overwrite = TRUE,
varsToDrop = c("device_id", "timestamp"))
i <- rxGetInfo(merged_new.xdf, numRows = 5, getVarInfo = TRUE)
i$data
## event_id longitude latitude new_id year month day hr min
## 1 11281 0.00 0.00 21595 116 4 Sa 11 36
## 2 12679 113.24 23.19 21595 116 4 Sa 12 18
## 3 49383 0.00 0.00 21595 116 4 Sa 15 44
## 4 131849 113.24 23.19 21595 116 4 Sa 12 6
## 5 205471 113.24 23.19 21595 116 4 F 15 36
We will now look at some summaries on this xdf file using rxSummary
and rxHistogram functions. Lets try to answer few questions to
understand the data better,
- How is the user activity spread across the week?
rxHistogram(~ day, merged.xdf)
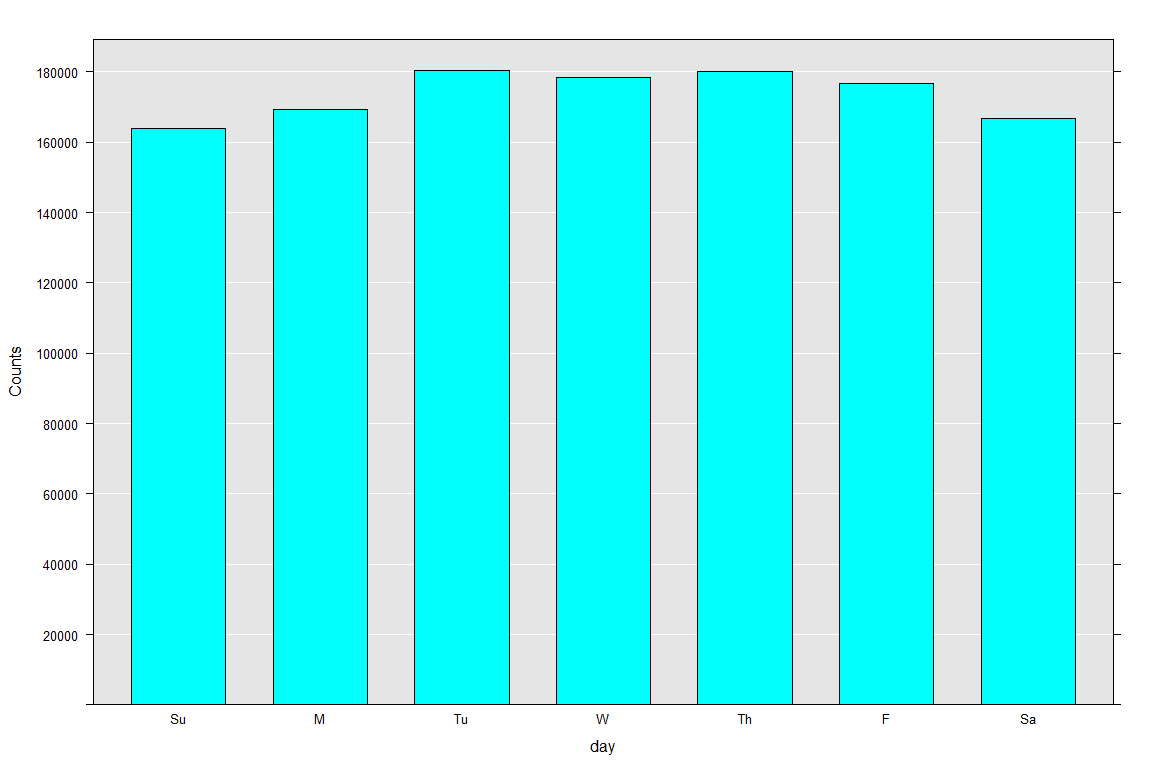
Surprisingly, the user activity is pretty uniform except for a minor bump on Tuesday and Thursday. The activity is the lowest during the weekends.
- How is the user activity spread across the day?
rxHistogram(~ F(hr), merged_new.xdf)
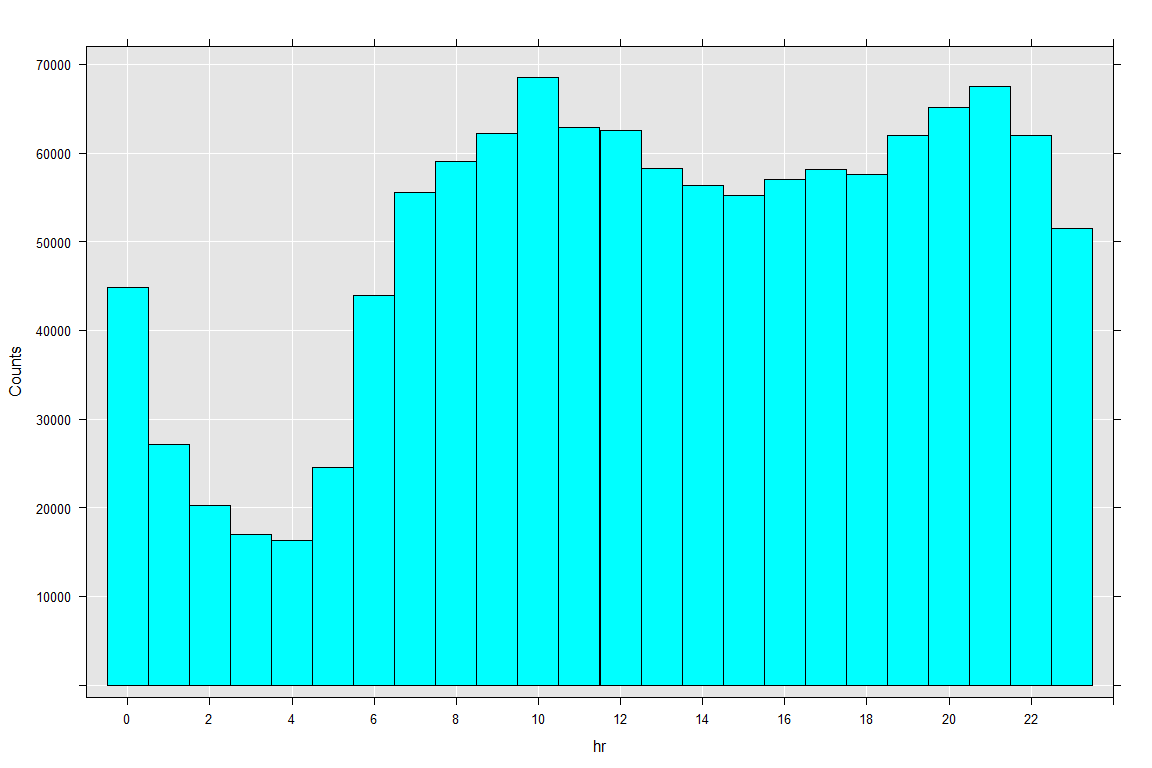
The user activity understandably is at its peak at 10AM and 9PM while at its lowest during the early hours of the day.
We will now move to the app_events.csv to extact the relevant
features.
app_events <- "app_events.csv"
app_events.xdf <- "app_events.xdf"
# Import the csv into an xdf data source
app_events.xdf <- rxImport(app_events,
app_events.xdf,
stringsAsFactors = FALSE,
overwrite = TRUE)
i <- rxGetInfo(app_events.xdf, getVarInfo = TRUE, numRows = 5)
i$numRows
## [1] 32473067
i$data
## event_id app_id is_installed is_active
## 1 2 5.927333e+18 1 1
## 2 2 -5.720079e+18 1 0
## 3 2 -1.633888e+18 1 0
## 4 2 -6.531843e+17 1 1
## 5 2 8.693964e+18 1 1
This file has about 32M rows with information about apps involved in
each and every event. We will merge this information with
merged_new.xdf.
all.merged.xdf <- rxMerge(merged_new.xdf,
app_events.xdf,
outFile = "all.merged.xdf",
type = "inner",
matchVars = "event_id",
overwrite = TRUE,
varsToDrop2 = c("is_installed", "is_active"))
i <- rxGetInfo(all.merged.xdf, getVarInfo = TRUE, numRows = 5)
i$data
## event_id longitude latitude new_id year month day hr min app_id
## 1 6 0 0 51155 116 4 Su 0 27 -7.377004e+18
## 2 6 0 0 51155 116 4 Su 0 27 6.284165e+18
## 3 6 0 0 51155 116 4 Su 0 27 5.927333e+18
## 4 6 0 0 51155 116 4 Su 0 27 3.433290e+18
## 5 6 0 0 51155 116 4 Su 0 27 -3.467200e+18
We finally managed to get all the data together with 12M rows. This is where Microsoft R server shines. It would have been impossible to get to this stage if we were conducting the experiment in an R Open environment. The parallel and distributed functions of MRS allow us to work with files residing on the disk, chunk them and perform transformations chunk wise in the memory. The final xdf file which is on the disk with 12M rows is only 21MB!
We can now continue aggregating this data to device level (new_id in
our case). To perform data aggregation we will take help of the
dplyrXdf
package.
# First install the package
devtools::install_github("RevolutionAnalytics/dplyrXdf")
library(dplyrXdf)
We will ignore year and month as we have data for only May 2016, so
that is constant across all devices.
agg.xdf <- all.merged.xdf %>%
group_by(new_id) %>%
summarise(med_lon = mean(longitude), # Mean of Longitude
med_lat = mean(latitude), # Mean of Latitude
num_apps = n(app_id)) %>% # Number of Apps
#gender = unique(gender),
#age = max(age),
#mode_day = names(which.max(table(day))),
#mode_hr = names(which.max(table(F(hr))))) %>%
persist("agg.xdf")
## Note: the dplyXdf does not provide support to arbitrary functions in summarise
# in its fastest form. Hence we will comment other variables for now and compute
# seperately.
i <- rxGetInfo(agg.xdf, getVarInfo = TRUE, numRows = 5)
i$data
## new_id med_lon med_lat num_apps
## 1 3 0.00000 0.00000 53
## 2 12 76.76741 20.38272 81
## 3 14 0.00000 0.00000 154
## 4 16 120.25431 31.89642 109
## 5 19 117.23261 39.12894 303
Step 6: Weekday with maximum activity (based on total number of apps)
To compute the maximum activity day for each device we perform the following steps:
- Compute total number of apps grouped by device and day
agg2.xdf <- all.merged.xdf %>%
group_by(new_id, day) %>%
summarise(num_apps = n(app_id)) %>%
persist("agg2.xdf")
i <- rxGetInfo(agg2.xdf, getVarInfo = TRUE, numRows = 5)
i$data
## new_id day num_apps
## 1 3 Su 53
## 2 16 Su 39
## 3 19 Su 99
## 4 30 Su 70
## 5 36 Su 147
- Select the MAX(
num_apps) grouped by device
agg3.xdf <- agg2.xdf %>%
group_by(new_id) %>%
summarise(max_num_apps = max(num_apps)) %>%
persist("agg3.xdf")
i <- rxGetInfo(agg3.xdf, getVarInfo = TRUE, numRows = 5)
i$data
## new_id max_num_apps
## 1 3 53
## 2 12 52
## 3 14 94
## 4 16 39
## 5 19 101
- Finally merge steps a & b. Compared to the capabilities of Open R this appears a bit awkward but the power lies in the parallel algorithms and spped of execution when working with big data files.
top_day.xdf <- rxMerge(agg3.xdf,
agg2.xdf,
outFile = "top_day.xdf",
type = "inner",
matchVars = c("new_id", "max_num_apps"),
overwrite = TRUE,
newVarNames2 = c(num_apps = "max_num_apps"))
# Select the first day in case there is a tie between 2 days.
# We can as well select last day
rxSort(inData = top_day.xdf,
outFile = top_day.xdf,
sortByVars = "new_id",
removeDupKeys = TRUE,
overwrite = TRUE)
i <- rxGetInfo(top_day.xdf, getVarInfo = TRUE, numRows = 5)
i$data
## new_id max_num_apps day
## 1 3 53 Su
## 2 12 52 W
## 3 14 94 Th
## 4 16 39 Su
## 5 19 101 F
Step 7: Day hour with maximum activity (based on total number of apps)
We will repeat the similar process to get the maximum activity hour for each device.
agg4.xdf <- all.merged.xdf %>%
group_by(new_id, hr) %>%
summarise(num_apps = n(app_id)) %>%
persist("agg4.xdf")
agg5.xdf <- agg4.xdf %>%
group_by(new_id) %>%
summarise(max_num_apps = max(num_apps)) %>%
persist("agg5.xdf")
top_hr.xdf <- rxMerge(agg5.xdf, agg4.xdf, outFile = "top_hr.xdf", type = "inner",
matchVars = c("new_id", "max_num_apps"),
overwrite = TRUE,
newVarNames2 = c(num_apps = "max_num_apps"))
# Select the first hour in case there is a tie between 2 different hours. We can as well select last hour.
rxSort(inData = top_hr.xdf,
outFile = top_hr.xdf,
sortByVars = "new_id",
removeDupKeys = TRUE,
overwrite = TRUE)
i <- rxGetInfo(top_hr.xdf, getVarInfo = TRUE, numRows = 5)
i$data
## new_id max_num_apps hr
## 1 3 53 14
## 2 12 52 14
## 3 14 40 22
## 4 16 46 12
## 5 19 103 17
Finally, merging top_day.xdf and top_hr.xdf with agg.xdf to bring
all features together
allfeat.xdf <- rxMerge(agg.xdf,
top_day.xdf,
outFile = "allfeat.xdf",
matchVars = "new_id",
type = "inner",
overwrite = TRUE
#varsToDrop2 = "max_num_apps",
#newVarNames2 = c(day = "top_day")
)
allfeat.xdf <- rxMerge(allfeat.xdf,
top_hr.xdf,
outFile = "allfeat.xdf",
matchVars = "new_id",
type = "inner",
overwrite = TRUE
#varsToDrop2 = "max_num_apps",
#newVarNames2 = c(hr = "top_hr")
)
i <- rxGetInfo(allfeat.xdf, getVarInfo = TRUE, numRows = 5)
i$data
## new_id med_lon med_lat num_apps max_num_apps.allfeat day
## 1 3 0.00000 0.00000 53 53 Su
## 2 12 76.76741 20.38272 81 52 W
## 3 14 0.00000 0.00000 154 94 Th
## 4 16 120.25431 31.89642 109 39 Su
## 5 19 117.23261 39.12894 303 101 F
## max_num_apps.top_hr hr
## 1 53 14
## 2 52 14
## 3 40 22
## 4 46 12
## 5 103 17
To conclude the feature engineering process we will map this information with demographic information and remove certain redundant columns.
allfeat.xdf <- rxMerge(gender_age,
allfeat.xdf,
outFile = "allfeat.xdf",
matchVars = "new_id",
type = "inner",
overwrite = TRUE,
varsToDrop2 = c("max_num_apps.allfeat", "max_num_apps.top_hr")
)
i <- rxGetInfo(allfeat.xdf, getVarInfo = TRUE, numRows = 5)
i$data
## gender age new_id med_lon med_lat num_apps day hr
## 1 M 35 3 0.00000 0.00000 53 Su 14
## 2 F 37 12 76.76741 20.38272 81 W 14
## 3 M 32 14 0.00000 0.00000 154 Th 22
## 4 F 28 16 120.25431 31.89642 109 Su 12
## 5 M 75 19 117.23261 39.12894 303 F 17
Note that we are ignoring the devices which do not have the app events related information.
We managed to get the data in to a good shape to explore if there is any relationship between the gender and max activity day and max activity hour.
Before we do that we need to convert the hr variable to factor as
well.
allfeatnew.xdf <- rxFactors(allfeat.xdf, outFile = "allfeatnew.xdf",
factorInfo = list(
hr = list(levels = 0:23,
newLevels = c( `0` = "0", `1` = "1",
`2` = "2", `3` = "3",
`4` = "4", `5` = "5",
`6` = "6", `7` = "7",
`8` = "8", `9` = "9",
`10` = "10", `11` = "11",
`12` = "12", `13` = "13",
`14` = "14", `15` = "15",
`16` = "16", `17` = "17",
`18` = "18", `19` = "19",
`20` = "20", `21` = "21",
`22` = "22", `23` = "23")
)),
overwrite = TRUE
)
i <- rxGetInfo(allfeatnew.xdf, getVarInfo = TRUE, numRows = 5)
i$varInfo
## Var 1: gender
## 2 factor levels: F M
## Var 2: age, Type: integer, Low/High: (11, 89)
## Var 3: new_id, Type: integer, Low/High: (3, 74645)
## Var 4: med_lon, Type: numeric, Low/High: (-169.4634, 136.1350)
## Var 5: med_lat, Type: numeric, Low/High: (-20.3239, 50.2403)
## Var 6: num_apps, Type: numeric, Low/High: (1.0000, 48132.0000)
## Var 7: day
## 7 factor levels: Su M Tu W Th F Sa
## Var 8: hr
## 24 factor levels: 0 1 2 3 4 ... 19 20 21 22 23
Data Exploration
Now that we have created new features, we can perform statistical test to ascertain the relation between the labels and independant variables (max activity day and hour).
Gender Vs Max Activity Day
Since both these variables are factors We can quickly perform a Chi-Squared to acces the good of fit.
ch1 <- rxCrossTabs(~ gender : day, data = allfeat.xdf)
rxChiSquaredTest(ch1)
## Chi-squared test of independence between gender and day
## X-squared df p-value
## 3.126907 6 0.7927584
The p-value is quite high for the relationship to be significant.
Gender Vs Max Activity Hour
We will perform the similar test between these 2 variables.
ch2 <- rxCrossTabs(~ gender : F(hr), data = allfeat.xdf)
rxChiSquaredTest(ch2)
## Chi-squared test of independence between gender and F_hr
## X-squared df p-value
## 162.7507 23 5.045395e-23
The low p-value indicates that the relationship is very strong and maximum activity hour can be a good predictor for the gender.
Gender Vs Age
Here Gender is a factor while Age is continuous variable. We can perform
t test using the
base R;s t.test function. But to perform this test we need to import
the data from xdf to data frame object as RevoScaleR does not offer
t-test function yet. We will write a helper function for that.
test <- function(var) {
df <- rxImport(allfeat.xdf, varsToKeep = c(var, "gender"))
print(t.test(as.formula(paste(var, "~", "gender")), data = df))
library(ggplot2)
df %>% ggplot(aes_string(x = "gender", y = var)) +
geom_boxplot() +
my_theme() +
labs(title = paste("gender Vs", var))
}
test("age")
##
## Welch Two Sample t-test
##
## data: age by gender
## t = 4.4002, df = 15029, p-value = 1.089e-05
## alternative hypothesis: true difference in means is not equal to 0
## 95 percent confidence interval:
## 0.3413012 0.8896352
## sample estimates:
## mean in group F mean in group M
## 32.63179 32.01633
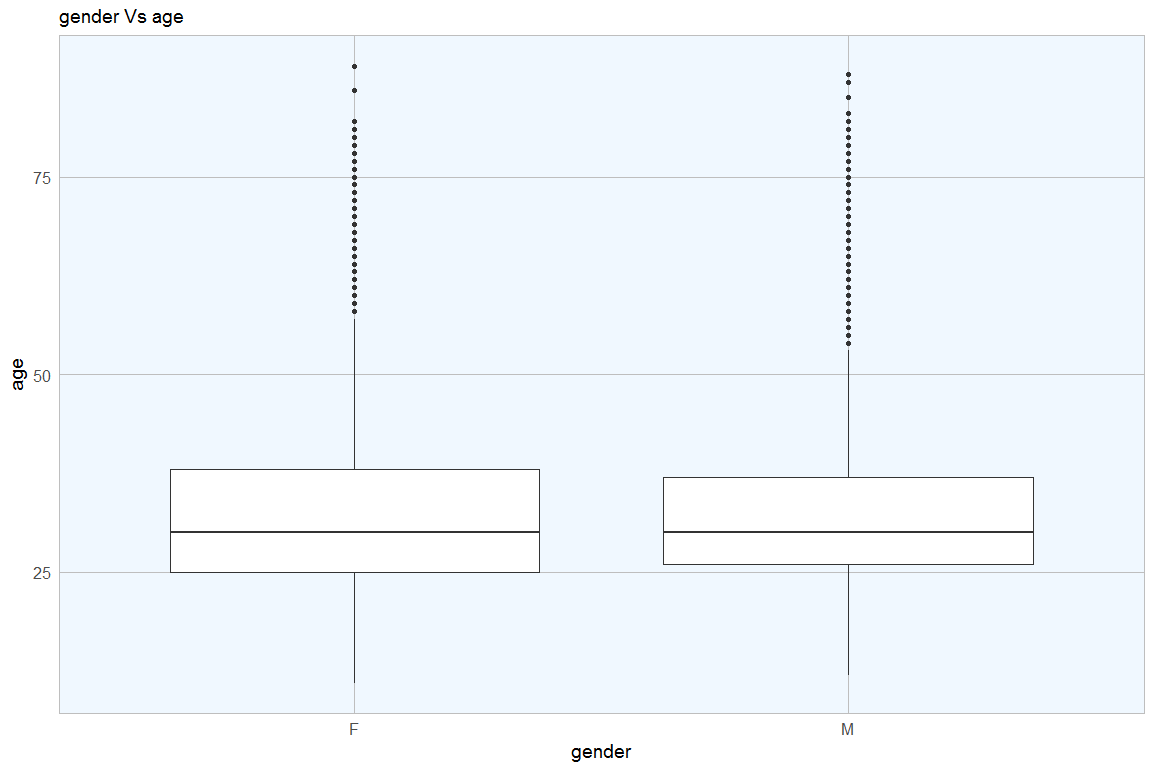 Again the p-value is quite low
indicating age can be a good indicator to predict gender.
Again the p-value is quite low
indicating age can be a good indicator to predict gender.
Next Step
In the following post we will start with this tidy data to build and evaluate various models to predict the gender. Please do leave a comment below or reach out to me for any query or if you find a bug. I will be happy to look into it.